Java JNI bindings for BWA(mem-lite)
Motivation
BWA 7.4(http://bio-bwa.sourceforge.net/) contains a small C example(https://github.com/lh3/bwa/blob/master/example.c) for running bwa-mem as a library (bwamem-lite). I created some JNI bindings to see if I can bind the C bwa library to java and get the same output than bwamem-lite. I put the code on github at https://github.com/lindenb/jbwa.
Example
(compare to https://github.com/lh3/bwa/blob/master/example.c )
System.loadLibrary("bwajni");
BwaIndex index=new BwaIndex(new File("hg19.fa"));
BwaMem mem=new BwaMem(index);
KSeq kseq=new KSeq(new File("input.fastq.gz");
ShortRead read=null;
while((read=kseq.next())!=null)
{
for(AlnRgn a: mem.align(read))
{
if(a.getSecondary()>=0) continue;
System.out.println( read.getName()+"\t"+ a.getStrand()+"\t"+ a.getChrom()+"\t"+
a.getPos()+"\t"+ a.getMQual()+"\t"+ a.getCigar()+"\t"+ a.getNm() );
}
}
kseq.dispose();
index.close();
mem.dispose();Testing
Here is the ouput of the JAVA version:
gunzip -c input.fastq.gz | head -n 4000 |\ java -Djava.library.path=src/main/native -cp src/main/java \ com.github.lindenb.jbwa.jni.Example human_g1k_v37.fasta -| tail HWI-1KL149:20:C1CU7ACXX:4:1101:3077:33410 + 3 38647538 60 89M11S 1 HWI-1KL149:20:C1CU7ACXX:4:1101:3396:33445 + 8 52567289 60 100M 1 HWI-1KL149:20:C1CU7ACXX:4:1101:10013:33288 - 1 156104115 60 100M 1 HWI-1KL149:20:C1CU7ACXX:4:1101:10390:33496 - 6 123824853 60 100M 1 HWI-1KL149:20:C1CU7ACXX:4:1101:13537:33483 + 2 157367092 60 100M 1 HWI-1KL149:20:C1CU7ACXX:4:1101:14139:33390 + 20 31413797 60 100M 1 HWI-1KL149:20:C1CU7ACXX:4:1101:14514:33458 + 2 179401813 60 100M 1 HWI-1KL149:20:C1CU7ACXX:4:1101:15292:33282 + 15 63335820 60 100M 1 HWI-1KL149:20:C1CU7ACXX:4:1101:16960:33276 - 12 110782784 60 100M 1 HWI-1KL149:20:C1CU7ACXX:4:1101:17355:33322 + 6 126077895 60 100M 1
And the ouput of the Native C version:
gunzip -c input.fastq.gz | head -n 4000 |\ bwa-0.7.4/bwamem-lite human_g1k_v37.fasta - | tail HWI-1KL149:20:C1CU7ACXX:4:1101:3077:33410 + 3 38647538 60 89M11S 1 HWI-1KL149:20:C1CU7ACXX:4:1101:3396:33445 + 8 52567289 60 100M 1 HWI-1KL149:20:C1CU7ACXX:4:1101:10013:33288 - 1 156104115 60 100M 1 HWI-1KL149:20:C1CU7ACXX:4:1101:10390:33496 - 6 123824853 60 100M 1 HWI-1KL149:20:C1CU7ACXX:4:1101:13537:33483 + 2 157367092 60 100M 1 HWI-1KL149:20:C1CU7ACXX:4:1101:14139:33390 + 20 31413797 60 100M 1 HWI-1KL149:20:C1CU7ACXX:4:1101:14514:33458 + 2 179401813 60 100M 1 HWI-1KL149:20:C1CU7ACXX:4:1101:15292:33282 + 15 63335820 60 100M 1 HWI-1KL149:20:C1CU7ACXX:4:1101:16960:33276 - 12 110782784 60 100M 1 HWI-1KL149:20:C1CU7ACXX:4:1101:17355:33322 + 6 126077895 60 100M 1
GUI
As a test I also created a swing-Based interface for BWA:
java -Djava.library.path=src/main/native -cp src/main/java \
com.github.lindenb.jbwa.jni.BwaFrame human_g1k_v37.fasta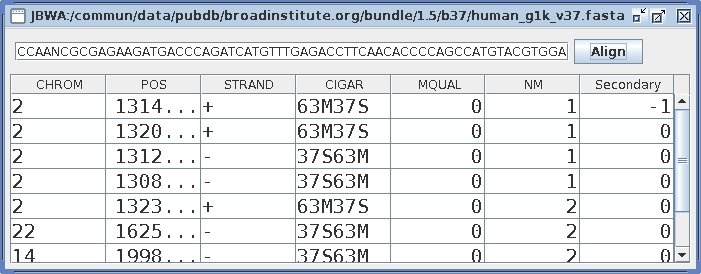
That's it,
Pierre