UCSC Genome Browser + SVG

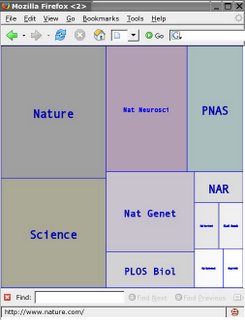
Pictures can be exported as a SVG file and edited with a SVG tool such as Inkscape or Adobe Illustrator
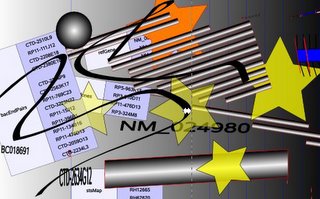
SVG is a vectorial format: Vector graphics editors allow to rotate, move, mirror, stretch, skew, generally perform affine transformations of objects, change z-order and combine the primitives into more complex objects.
Updated 2010-08-12: source code
/*
author:
- Pierre Lindenbaum PhD plindenbaum (at) yahoo (dot) fr http://www.integragen.com
Permission is hereby granted, free of charge, to any person obtaining
a copy of this software and associated documentation files (the
``Software''), to deal in the Software without restriction, including
without limitation the rights to use, copy, modify, merge, publish,
distribute, sublicense, and/or sell copies of the Software, and to
permit persons to whom the Software is furnished to do so, subject to
the following conditions:
The above copyright notice and this permission notice shall be included
in all copies or substantial portions of the Software.
The name of the authors when specified in the source files shall be
kept unmodified.
THE SOFTWARE IS PROVIDED ``AS IS'', WITHOUT WARRANTY OF ANY KIND, EXPRESS
OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF
MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT.
IN NO EVENT SHALL 4XT.ORG BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE,
ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE
USE OR OTHER DEALINGS IN THE SOFTWARE.
$Id: $
$Author: $
$Revision: $
$Date: $
$Locker: $
$RCSfile: $
$Source: $
$State: $
$Name: $
$Log: $
*************************************************************************/
$sqllimitnumber="50";
$sqllimit=" limit ".$sqllimitnumber;
$fontsize="14";
$chrs= array(
"chr1","chr2","chr3","chr4","chr5","chr6","chr7","chr8","chr9","chr10",
"chr11","chr12","chr13","chr14","chr15","chr16","chr17","chr18","chr19","chr20",
"chr21","chr22","chrX","chrY"
);
$tables= array(
"knownGene"=>"full",
"all_mrna"=>"packed",
"refGene"=>"full",
"bacEndPairs"=>"full",
"fishClones"=>"full",
"stsMap"=>"full",
"snp"=>"packed"
);
$viewAs= array("full","packed","hide");
/**
*
* Item an item on the genome
*
*/
class Item
{
var $name;
var $track;
//constructor
function Item(&$track)
{
$this->track=$track;
}
//get 5' bound
function getStart()
{
return -1;
}
//get 3' bound
function getEnd()
{
return -1;
}
//returns wether 2 Item overlap
function overlap($other)
{
return (!($this->getEnd()<$other->getStart() || $other->getEnd()<$this->getStart()));
}
//returns wether 2 Item overlap on screen
function overlapOnScreen($other)
{
$start1= $this->base2pixel(min($this->getStart(),$this->getEnd()));
$end1= 1+$this->base2pixel(max($this->getStart(),$this->getEnd()));
$start2= $other->base2pixel(min($other->getStart(),$other->getEnd()));
$end2= 1+$other->base2pixel(max($other->getStart(),$other->getEnd()));
return (!($end1 < $start2 || $end2 < $start1));
}
//return a URL for this item
function getURL()
{
return "http://www.ncbi.nlm.nih.gov/gquery/gquery.fcgi?term=".htmlentities($this->name);
}
//convert a base position to the screen
function base2pixel($base)
{
$left= $this->track->browser->leftMarginWidth();
$screenw= $this->track->browser->genomeWidth();
return $left+$screenw* (($base - $this->track->browser->start)/($this->track->browser->end - $this->track->browser->start));
}
//force a pixel to be in the drawing area
function trimPixel($pix)
{
$pix= min($this->track->browser->getWidth(),$pix);
$pix= max($this->track->browser->leftMarginWidth(),$pix);
return $pix;
}
//return the height of this item on the screen
function getHeight()
{
return $this->track->browser->featureHeight;
}
//writes symbol for orientation
function writeStrand($out,$pixx1,$pixx2,$midy)
{
if($this->strand=="?") return;
//write orientation
for($i=$pixx1; $i<= $pixx2;$i+=10)
{
fwrite($out,"<svg:use x='".$i."' y='".$midy."' xlink:href='#".($this->strand=='-'?"minus":"plus")."'/>");
}
}
//write a svg:a link containg $svg
function writeAnchor($out,$svg)
{
fwrite($out,"<svg:a xlink:href='".$this->getURL()."' xlink:title='".htmlentities($this->name)."'>".
$svg."</svg:a>"
);
}
}
/**
*
* SimpleItem such as knownGene
*
*/
class SimpleItem extends Item
{
var $chromStart;
var $chromEnd;
var $strand;
//constructor
function SimpleItem(&$track)
{
parent::Item( $track);
$this->strand="?";
}
//get 5' bound
function getStart()
{
return $this->chromStart;
}
//get 3' bound
function getEnd()
{
return $this->chromEnd;
}
//write as SVG
function toSVG($out,$y,$isPacked=TRUE)
{
fwrite($out,"<svg:g>\n");
if($isPacked==FALSE)
{
$this->writeAnchor($out,"<svg:text x='".($this->track->browser->trackMarginWidth()+($this->track->browser->itemMarginWidth()/2.0))."' y='".($y+$this->getHeight()-$GLOBALS["fontsize"]/2.0)."' text-anchor='middle'>".htmlentities($this->name)."</svg:text>");
}
$pixx1= $this->trimPixel($this->base2pixel($this->chromStart));
$pixx2= $this->trimPixel($this->base2pixel($this->chromEnd));
$midy= ($y+$this->getHeight()/2.0);
//write orientation
$this->writeStrand($out,$pixx1,$pixx2,$midy);
$h2= $this->getHeight()/5.0;
$svg = "<svg:rect x='".$pixx1."' y='".($midy-$h2)."' width='".($pixx2-$pixx1)."' height='".($h2*2)."' style='stroke:red;fill:url(#metal);'/>";
if($isPacked)
{
$this->writeAnchor($out,$svg);
}
else
{
fwrite($out,$svg);
}
fwrite($out,"</svg:g>");
}
}
/**
*
* BEDItem such as knownGene
*
*/
class BEDItem extends Item
{
var $txStart;
var $txEnd;
var $cdsStart;
var $cdsEnd;
var $exonCount;
var $exonStarts;
var $exonEnds;
//constructor
function BEDItem(&$track)
{
parent::Item($track);
}
//5' bound
function getStart()
{
return $this->txStart;
}
//3' bound
function getEnd()
{
return $this->txEnd;
}
//write as SVG
function toSVG($out,$y,$isPacked=TRUE)
{
fwrite($out,"<svg:g>");
if($isPacked==FALSE)
{
$this->writeAnchor($out,"<svg:text x='".($this->track->browser->trackMarginWidth()+($this->track->browser->itemMarginWidth()/2.0))."' y='".($y+$this->getHeight()-$GLOBALS["fontsize"]/2.0)."' text-anchor='middle'>".htmlentities($this->name)."</svg:text>"
);
}
$pixx1= $this->trimPixel($this->base2pixel($this->txStart));
$pixx2= $this->trimPixel($this->base2pixel($this->txEnd));
$midy= ($y+$this->getHeight()/2.0);
$this->writeStrand($out,$pixx1,$pixx2,$midy);
//write gene
fwrite($out,"<svg:line x1='".$pixx1."' y1='".$midy."' x2='".$pixx2."' y2='".$midy."' stroke='black'/>");
$pixx1= $this->trimPixel($this->base2pixel($this->cdsStart));
$pixx2= $this->trimPixel($this->base2pixel($this->cdsEnd));
//write mRNA
$h2= $this->getHeight()/17.0;
fwrite($out,"<svg:rect x='".$pixx1."' y='".($midy-$h2)."' width='".($pixx2-$pixx1)."' height='".($h2*2)."' stroke='black' fill='blue'/>");
//write translation
$h2= $this->getHeight()/6.0;
for($i=0;$i< $this->exonCount;++$i)
{
$pixx1= $this->trimPixel($this->base2pixel($this->exonStarts[$i]));
$pixx2= $this->trimPixel($this->base2pixel($this->exonEnds[$i]));
$svg="<svg:rect x='".$pixx1."' y='".($midy-$h2)."' width='".($pixx2-$pixx1)."' height='".($h2*2)."' style='stroke:red;fill:url(#metal);'/>";
if($isPacked==FALSE)
{
fwrite($out,$svg);
}
else
{
$this->writeAnchor($out,$svg);
}
}
fwrite($out,"</svg:g>");
}
}
/**
*
* AlignItem such as all_mrna
*
*/
class AlignItem extends Item
{
var $tStart;
var $tEnd;
var $blockCount;
var $blockSizes;
var $tStarts;
//constructor
function AlignItem(&$track)
{
parent::Item($track);
}
//5' bound
function getStart()
{
return $this->tStart;
}
//3' bound
function getEnd()
{
return $this->tEnd;
}
//write as SVG
function toSVG($out,$y,$isPacked=TRUE)
{
fwrite($out,"<svg:g>");
if($isPacked==FALSE)
{
$this->writeAnchor($out,"<svg:text x='".($this->track->browser->trackMarginWidth()+($this->track->browser->itemMarginWidth()/2.0))."' y='".($y+$this->getHeight()-$GLOBALS["fontsize"]/2.0)."' text-anchor='middle'>".htmlentities($this->name)."</svg:text>"
);
}
$pixx1= $this->trimPixel($this->base2pixel($this->getStart()));
$pixx2= $this->trimPixel($this->base2pixel($this->getEnd()));
$midy= ($y+$this->getHeight()/2.0);
$this->writeStrand($out,$pixx1,$pixx2,$midy);
//write gene
fwrite($out,"<svg:line x1='".$pixx1."' y1='".$midy."' x2='".$pixx2."' y2='".$midy."' stroke='black'/>");
//write blocks
$h2= $this->getHeight()/6.0;
for($i=0;$i< $this->blockCount;++$i)
{
$pixx1= $this->trimPixel($this->base2pixel($this->tStarts[$i]));
$pixx2= $this->trimPixel($this->base2pixel($this->tStarts[$i]+$this->blockSizes[$i]));
$svg="<svg:rect x='".$pixx1."' y='".($midy-$h2)."' width='".($pixx2-$pixx1)."' height='".($h2*2)."' style='stroke:red;fill:url(#metal);'/>";
if($isPacked==FALSE)
{
fwrite($out,$svg);
}
else
{
$this->writeAnchor($out,$svg);
}
}
fwrite($out,"</svg:g>");
}
}
/**
*
* class Track
* a track is a vector of Item
*
*/
class Track
{
var $name;
var $url;
var $browser;
var $items;
var $index2row;
var $nRows;
//constructor
function Track(&$browser,$name,$url)
{
$this->name = $name;
$this->url = $url;
$this->browser= $browser;
$this->items = array();
$this->index2row = null;
$this->nRows=-1;
}
//number of items
function getItemCount()
{
return count($this->items);
}
//add a new item in the track
function add($item)
{
$this->items[ $this->getItemCount() ] = $item;
$this->nRows=-1;
}
//return width on screen
function getWidth()
{
return $this->browser->getWidth();
}
//return height on screen
function getHeight()
{
if($this->isPacked())
{
return $this->nRows* $this->browser->featureHeight;
}
else
{
$h=0;
foreach($this->items as $K=>$V)
{
$h += $V->getHeight();
}
return $h;
}
}
//return wether is track has been packed
function isPacked()
{
return $this->nRows!=-1;
}
//pack this track
function packTrack()
{
$count= $this->getItemCount();
$this->index2row= array();
$this->nRows=1;
$this->index2row[0]=0;
for($i=1;$i< $count;$i++)
{
$itemi = $this->items[$i];
$choosenRow=0;
$done=FALSE;
while($done==FALSE)
{
$done=TRUE;
for($j=0;$j<$i;$j++)
{
if($this->index2row[$j]!=$choosenRow) continue;
$itemj = $this->items[$j];
if($itemi->overlapOnScreen($itemj)==TRUE)
{
$choosenRow++;
$done=FALSE;
break;
}
}
if($choosenRow>=$this->nRows)
{
$this->nRows++;
break;
}
}
$this->index2row[$i]=$choosenRow;
}
}
//write as SVG
function toSVG($out,$y)
{
$width = $this->getWidth();
fwrite($out,"<svg:g id=\"".$this->name."\">\n");
//track name
fwrite($out, "<svg:g>".
"<svg:a xlink:href='".$this->url."' xlink:title='".$this->name."'>".
"<svg:rect x='0' y='".$y."' width='".($this->browser->trackMarginWidth()).
"' height='".$this->getHeight()."' fill='rgb(240,240,255)'/>".
"</svg:a>".
"<svg:text x='0' y='0' text-anchor='middle' transform='translate(".(($this->browser->trackMarginWidth())/2.0).",".($y+$GLOBALS["fontsize"]/2.0+($this->getHeight())/2.0).") rotate(0)'>".$this->name."</svg:text>".
"</svg:g>\n"
);
fwrite($out,"<svg:rect x='".($this->browser->trackMarginWidth())."' y='".$y."' width='".($this->browser->itemMarginWidth()).
"' height='".$this->getHeight()."' fill='rgb(200,200,255)'/>\n");
if($this->isPacked()==TRUE)
{
$count= $this->getItemCount();
for($i=0;$i< $this->nRows;$i++)
{
for($j=0;$j<$count;$j++)
{
if($this->index2row[$j]!=$i) continue;
$this->items[$j]->toSVG($out,$y,TRUE);
}
$y+=$this->browser->featureHeight;
}
}
else
{
foreach($this->items as $K=>$V)
{
$V->toSVG($out,$y,FALSE);
$y+=$this->browser->featureHeight;
}
}
fwrite($out,"</svg:g>");
}
}
/**************
*
* Browser
* a browser is a vector of tracks
*
*/
class Browser
{
var $build;
var $chrom;
var $tracks;
var $start;
var $end;
var $featureHeight;
//constructor
function Browser($build,$chrom,$start,$end)
{
$this->build = $buid;
$this->chrom = $chrom;
$this->start = $start;
$this->end = $end;
$this->featureHeight=24;
$this->tracks=array();
}
//number of tracks
function getTrackCount()
{
return count( $this->tracks);
}
//add a new track
function add(&$track)
{
$this->tracks[ $this->getTrackCount() ]=& $track;
}
//width on screen
function getWidth()
{
return 800;
}
//height on screen
function getHeight()
{
$h=0;
foreach($this->tracks as $key=>$value)
{
$h+= $value->getHeight();
}
return $h;
}
//margin width for labels
function itemMarginWidth()
{
return $this->getWidth()/6.0;
}
//margin width for track
function trackMarginWidth()
{
return $this->getWidth()/6.0;
}
//left margin width = sum(item+track)
function leftMarginWidth()
{
return $this->trackMarginWidth()+$this->itemMarginWidth();
}
//drawing area
function genomeWidth()
{
return $this->getWidth()-$this->leftMarginWidth();
}
//pack the named track
function packTrack($name)
{
for($i=0;$i< count($this->tracks);$i++)
{
if($this->tracks[$i]->name==$name)
{
$this->tracks[$i]->packTrack();
}
}
}
//write as SVG
function toSVG($out)
{
if($out==NULL)
{
$out=fopen("php://output","w")or die ("stdout?");
}
$height=$this->getHeight();
$fh2 = ($this->featureHeight)/5.0;
fwrite($out, "<svg:svg xmlns:svg='http://www.w3.org/2000/svg' xmlns:xlink='http://www.w3.org/1999/xlink' width='".$this->getWidth()."' height='".$height."' stroke='black' stroke-width='0.5' font-size='".$GLOBALS["fontsize"]."'>".
"<svg:title>".$this->chrom.":".$this->start."-".$this->end."</svg:title>".
"<svg:desc>Genome Browser with SVG Pierre Lindenbaum plindenbaum (@) yahoo (dot) fr </svg:desc>".
"<svg:defs>".
"<svg:polyline id='plus' stroke='black' fill='none' points='".
"-".$fh2.",-".$fh2." ".
" 0,0 ".
"-".$fh2.",".$fh2."'/>".
"<svg:polyline id='minus' stroke='black' fill='none' points='".
"".$fh2.",-".$fh2." ".
" 0,0 ".
"".$fh2.",".$fh2."'/>".
"<svg:linearGradient x1='0%' y1='0%' x2='0%' y2='100%' id='metal'>\n".
"<svg:stop offset=\"5%\" stop-color=\"black\"/>\n".
"<svg:stop offset=\"50%\" stop-color=\"whitesmoke\"/>\n".
"<svg:stop offset=\"95%\" stop-color=\"black\"/>\n".
"</svg:linearGradient>\n".
"</svg:defs>"
);
fwrite($out,"<svg:rect x='0' y='0' ".
"width='".$this->getWidth()."' height='".$height."' stroke='blue' fill='white' ".
"/>"
);
//write vertical bar
fwrite($out,"<svg:g>");
for($i=1;$i<10;$i++)
{
$x= $this->leftMarginWidth()+($this->genomeWidth()/10.0)*$i;
fwrite($out,"<svg:line x1='".$x."' y1='0' x2='".$x."' y2='".$height."' stroke='blue' />");
}
fwrite($out,"</svg:g>");
$y=0;
foreach($this->tracks as $key=>$value)
{
$value->toSVG($out,$y);
$y+= $value->getHeight();
}
fwrite($out,"<svg:rect x='0' y='0' ".
"width='".$this->getWidth()."' height='".$height."' stroke='blue' fill='none' ".
"/>"
);
fwrite($out, "</svg:svg>");
}
}
/** return a POST parameter, convenient method that can be changed to __GET */
function getParameter($s)
{
return $_POST[$s];
}
/* performs a query in gene tracks */
function doGeneQuery($con,&$browser,$build,$trackname)
{
$prompt="select ".
"name,strand,txStart,txEnd,cdsStart,cdsEnd,exonCount,exonStarts,exonEnds".
" from ".
" ".$build.".".$trackname.
" where ".
" chrom=\"".mysql_escape_string($browser->chrom) ."\" and not(".
" txEnd < \"".mysql_escape_string($browser->start)."\" or \"".
mysql_escape_string($browser->end)."\" < txStart ".
") ".$GLOBALS["sqllimit"];
//echo "<!-- ".$prompt." -->";
$result = mysql_query($prompt,$con);
if(!$result)
{
echo "<!-- Bad query ".$prompt." -->";
}
else
{
$track= new Track($browser,$trackname,"http://www.genome.ucsc.edu/cgi-bin/hgTrackUi?g=".$trackname);
while($row=mysql_fetch_array($result))
{
$item= new BEDItem(&$track);
$item->name= $row[0];
$item->strand= $row[1];
$item->txStart = $row[2];
$item->txEnd = $row[3];
$item->cdsStart = $row[4];
$item->cdsEnd = $row[5];
$item->exonCount= $row[6];
$item->exonStarts= split(",",$row[7]);
$item->exonEnds= split(",",$row[8]);
$track->add( $item);
}
$browser->add(& $track);
}
}
/* performs a generic query */
function doSimpleQuery($con,&$browser,$build,$trackname,$N,$T,$C,$S,$E)
{
$prompt="select ".
"$N,".($T==NULL?"\"?\"":$T).",$S,$E ".
" from ".
" ".$build.".".$trackname.
" where ".
" $C=\"".mysql_escape_string($browser->chrom) ."\" and not(".
" $E < \"".mysql_escape_string($browser->start)."\" or \"".
mysql_escape_string($browser->end)."\" < $S ".
") ".$GLOBALS["sqllimit"];
//echo "<!-- ".$prompt." -->";
$result = mysql_query($prompt,$con);
if(!$result)
{
echo "<!-- Bad query ".$prompt." -->";
}
else
{
//echo "<!-- query ".$prompt." -->";
$track= new Track($browser,$trackname,"http://www.genome.ucsc.edu/cgi-bin/hgTrackUi?g=".$trackname);
while($row=mysql_fetch_array($result))
{
$item= new SimpleItem(&$track);
$item->name= $row[0];
$item->strand= ($T==NULL?"?":$row[1]);
$item->chromStart = $row[2];
$item->chromEnd = $row[3];
$track->add( $item);
}
$browser->add(& $track);
}
}
/* performs a alignment query */
function doAlignQuery($con,&$browser,$build,$trackname,$N,$T,$C,$S,$E,$BC,$BS,$BL)
{
$prompt="select ".
"$N,".($T==NULL?"\"?\"":$T).",$S,$E,$BC,$BS,$BL".
" from ".
" ".$build.".".$trackname.
" where ".
" $C=\"".mysql_escape_string($browser->chrom) ."\" and not(".
" $E < \"".mysql_escape_string($browser->start)."\" or \"".
mysql_escape_string($browser->end)."\" < $E ".
") ".$GLOBALS["sqllimit"];
//echo "<!-- ".$prompt." -->";
$result = mysql_query($prompt,$con);
if(!$result)
{
echo "<!-- Bad query ".$prompt." -->";
}
else
{
$track= new Track($browser,$trackname,"http://www.genome.ucsc.edu/cgi-bin/hgTrackUi?g=".$trackname);
while($row=mysql_fetch_array($result))
{
$item= new AlignItem(&$track);
$item->name= $row[0];
$item->strand= $row[1];
$item->tStart = $row[2];
$item->tEnd = $row[3];
$item->blockCount= $row[4];
$item->tStarts= split(",",$row[5]);
$item->blockSizes= split(",",$row[6]);
$track->add( $item);
}
$browser->add(& $track);
}
}
$svgonly= getParameter("svgonly");
//$build= getParameter("build"); you can un-comment this
if(!isset($build)) $build="hg17";
if(isset($build)) $build=trim($build);
$chrom= getParameter("chrom");
if(isset($chrom)) $chrom=trim($chrom);
$start= getParameter("start");
if(isset($start)) $start=intval($start);
$end= getParameter("end");
if(isset($end)) $end=intval($end);
if(
isset($chrom) &&
isset($start) &&
isset($end) &&
$start<$end
)
{
$title=$chrom.":".$start."-".$end;
}
else
{
$title=NULL;
}
if($title==NULL || isset($svgonly)==FALSE)
{
header("Content-type: application/xhtml+xml");
?>
<!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.1//EN" "http://www.w3.org/TR/xhtml11/DTD/xhtml11.dtd">
<html xmlns="http://www.w3.org/1999/xhtml" xml:lang="en">
<head>
<link rel="alternate" type="application/rss+xml" href="../rss/rss.txt"/>
<meta name="dc.description" content="UCSC Genome Browser with SVG"/>
<meta name="dc.keywords" content="UCSC; Genome Browser; goldenpath; SVG; PHP;mozilla; firefox; genomics; bioinformatics; integragen"/>
<meta name="dc.author" content="Pierre Lindenbaum"/>
<title><?php echo ($title==NULL?"UCSC Genome Browser with SVG":$title); ?></title>
</head>
<body>
<h1>Displaying Data from the UCSC Genome Browser With SVG</h1>
<h4>Pierre Lindenbaum PhD, 2006</h4>
<div align="center" style="font-size:9pt; background-color:#DDDDDD;border-color:black; border-width:1px; ; border-style:solid;"><br/>
<?php
if(isset($sqllimitnumber))
{
echo "<br/>For security reason, the number of items per track is limited to <b style='color:red;'>$sqllimitnumber</b>.<br/><br/>";
}
?>
<form method="POST">
<input type="hidden" name="build" value="hg17" />
Assembly:<span style='border-color:gray; border-width:1px; ; border-style:dashed;background-color:white;'><?php echo $build; ?></span>
<label for="chrom">Chromosome:</label><select name="chrom" id="chrom">
<?php
echo "<input type=\"hidden\" name=\"_rand\" value=\"".time()."\"/>";
foreach($chrs as $K)
{
echo "<option value=\"".$K."\"";
if(isset($chrom) && $K==$chrom) echo " selected=\"true\" style='background-color:#EE00EE'";
echo ">".$K."</option>";
}
?>
</select>
<label for="start">Start:</label><input type="text" id="start" name="start" value="<?php echo (isset($start)?$start:910000); ?>"/>
<label for="end">End:</label><input type="text" id="end" name="end" value="<?php echo (isset($end)?$end:930000); ?>"/>
<?php
echo "<br/><table><tr>";
foreach($tables as $T=>$D)
{
$v=getParameter($T);
if(isset($v) && in_array($v,$viewAs))
{
$D=$v;
$tables[$T]=$D;
}
echo "<th><label for='$T'>$T</label></th><td><select id='$T' name='$T'>";
foreach($viewAs as $V)
{
echo "<option value='$V'";
if($V==$D) echo " selected=\"true\" style='background-color:#EE00EE' ";
echo ">".$V."</option>";
}
echo "</select></td>\n";
}
echo "</tr></table>";
?>
<input type="submit" name="Submit" value='Display as XHTML'/>
<input type="submit" name="svgonly" value='Save As SVG' />
<br/><br/></form>
</div>
<p/>
<div align="center" style="font-size:9pt; background-color:#DDDDFF;border-color:#DDDDDD; border-width:1px; ; border-style:solid;"><br/>
<?php
}
else
{
header("Content-disposition: attachment; filename=\"".$chrom."_".$start."_".$end."\"");
header("Content-type: image/svg+xml");
?>
<!DOCTYPE svg PUBLIC "-//W3C//DTD SVG 1.0//EN" "http://www.w3.org/TR/2001/REC-SVG-20010904/DTD/svg10.dtd">
<?php
}
if(
isset($chrom) &&
isset($start) &&
isset($end) &&
$start<$end
)
{
if(isset($svgonly)==FALSE)
{
echo "<a style='font-size:200%;' href='http://www.genome.ucsc.edu/cgi-bin/hgTracks?clade=vertebrate&org=Human&db=$build&position=$chrom%3A$start-$end&pix=1000'>($build)$title</a><br/>";
}
$con = mysql_connect("genome-mysql.cse.ucsc.edu", "genome");
if(!$con)
{
die("Cannot connect :=".mysql_error($con));
}
else
{
$browser= new Browser($build,$chrom,$start,$end);
if($tables["knownGene"]!="hide")
{
doGeneQuery($con,&$browser,$build,"knownGene");
if($tables["knownGene"]=="packed") $browser->packTrack("knownGene");
}
if($tables["refGene"]!="hide")
{
doGeneQuery($con,&$browser,$build,"refGene");
if($tables["refGene"]=="packed") $browser->packTrack("refGene");
}
if($tables["all_mrna"]!="hide")
{
doAlignQuery($con,&$browser,$build,"all_mrna","qName","strand","tName","tStart","tEnd","blockCount","tStarts","blockSizes");
if($tables["all_mrna"]=="packed") $browser->packTrack("all_mrna");
}
if($tables["bacEndPairs"]!="hide")
{
doSimpleQuery($con,&$browser,$build,"bacEndPairs","name","strand","chrom","chromStart","chromEnd");
if($tables["bacEndPairs"]=="packed") $browser->packTrack("bacEndPairs");
}
if($tables["fishClones"]!="hide")
{
doSimpleQuery($con,&$browser,$build,"fishClones","name",NULL,"chrom","chromStart","chromEnd");
if($tables["fishClones"]=="packed") $browser->packTrack("fishClones");
}
if($tables["stsMap"]!="hide")
{
doSimpleQuery($con,&$browser,$build,"stsMap","name",NULL,"chrom","chromStart","chromEnd");
if($tables["stsMap"]=="packed") $browser->packTrack("stsMap");
}
if($tables["snp"]!="hide")
{
doSimpleQuery($con,&$browser,$build,"snp125","name","strand","chrom","chromStart","chromEnd");
if($tables["snp"]=="packed") $browser->packTrack("snp");
}
$browser->toSVG(NULL);
mysql_close($con);
}
}
if($title==NULL || isset($svgonly)==FALSE)
{
if($title==NULL) {
?>
<div align="left">This <a href="http://www.php.net">PHP</a> script display tracks from the <a href="http://www.genome.ucsc.edu/">UCSC Genome Browser</a> using <a href="http://www.w3.org/Graphics/SVG/">SVG</a> and the <a href="http://genome.ucsc.edu/FAQ/FAQdownloads#download29">public mysql connection to their database</a>. As <a
href="http://www.mozilla.org">Firefox</a> <a href="http://developer.mozilla.org/en/docs/SVG_in_Firefox_1.5"> now supports the SVG</a> format, this drawing can be displayed in your
web browser.</div>
<br/>
<p style="font-size:200%;"><u>Must</u> be viewed with <u><b>Firefox 1.5 or higher</b></u> : <a href="http://www.spreadfirefox.com/?q=affiliates&id=0&t=45"><img alt="Get Firefox!" title="Get Firefox!" src="http://sfx-images.mozilla.org/affiliates/Buttons/80x15/blue_1.gif" border="0"/></a></p>
<br/>
Pictures can be exported as a SVG file and edited with a SVG tool such as <a href="http://www.inkscape.org/" target="inkscape" >Inkscape</a> or <a target="illustrator" href="http://www.adobe.com/svg/tools.html">Adobe Illustrator</a>
<img src="01ucsc2svgInkscape.jpeg" alt="01ucsc2svgInkscape.jpeg"/><br/>
<br/>
SVG is a <a href="http://en.wikipedia.org/wiki/Vector_graphics">vectorial format</a>: <cite>Vector graphics editors allow to rotate, move, mirror, stretch, skew, generally perform affine transformations of objects, change z-order and combine the primitives into more complex objects.</cite><br/>
<img src="02ucsc2svgInkscape.jpeg" alt="02ucsc2svgInkscape.jpeg"/>
<br/>
<?php
echo "<div align='left'><h3>The PHP Code</h3><pre style='background-color:lightgray'>";
echo htmlspecialchars(file_get_contents("ucsc.php"));
echo "</pre></div>";
}
?>
</div>
<h3>How to cite this code?</h3>
<p>Displaying data from the UCSC GenomeBrowser using SVG: Pierre Lindenbaum 2006. Integragen</p>
<p><a href="http://www.genome.ucsc.edu/">UCSC GenomeBrowser</a>: The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006 Jan 1;34(Database issue):D590-8. PMID: <a href="http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=16381938">16381938</a></p>
<h3>Links</h3>
<ul>
<li><a href="http://www.genome.ucsc.edu/">UCSC GenomeBrowser</a>: The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006 Jan 1;34(Database issue):D590-8. PMID: <a href="http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=16381938">16381938</a></li>
<li><a href="http://www.integragen.com">Integragen</a></li>
<li><a href="http://www.urbigene.com">Home</a></li>
<li><a href="http://plindenbaum.blogspot.com">blog</a></li>
</ul>
<hr/>
<adress>
<a href="http://plindenbaum.blogspot.com">Pierre Lindenbaum PhD</a><br/>
lindenb ( at ) integragen (dot) com<br/>
<a href="http://www.integragen.com">Integragen</a><br/>
4, rue Pierre Fontaine
91000 EVRY.
</adress>
<div align="center"><a href="http://www.integragen.com"><img src="http://www.integragen.com/img//title.png"
border="1" /></a></div>
<script src="http://www.google-analytics.com/urchin.js" type="text/javascript">
</script>
<script type="text/javascript">
_uacct = "UA-XXXXX-2";
urchinTracker();
</script>
</body>
</html>
<?php
}