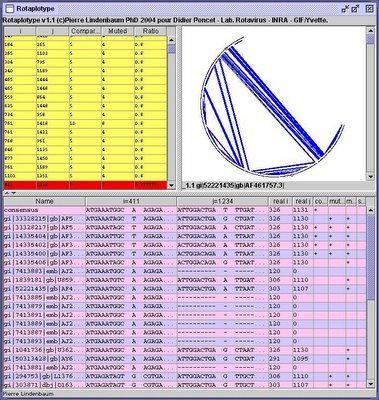
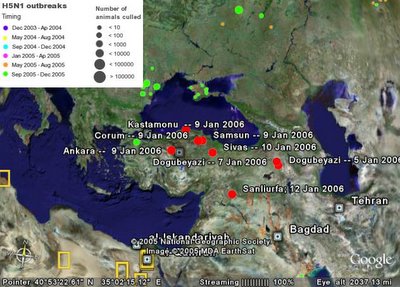
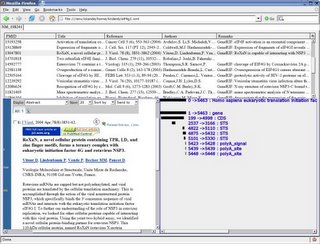
I still had problems with the XUL layout and I was not able to display more than one tabpanel (the others were frozen). But creating such image on the fly could be great way to display interaction genomic maps. As an example, the
(aka golden path) might use this system to display its tracks and make it interactive.
<?xml version="1.0" ?>
<xsl:stylesheet version="1.0"
xmlns:xsl="
http://www.w3.org/1999/XSL/Transform"
xmlns:svg="
http://www.w3.org/2000/svg"
xmlns:xlink="
http://www.w3.org/1999/xlink"
xmlns:xul="
http://www.mozilla.org/keymaster/gatekeeper/there.is.only.xul"
xmlns:h="
http://www.w3.org/TR/REC-html40"
>
<!--
author : Pierre Lindenbaum PhD
blog:
http://plindenbaum.blogspot.com mail: plindenbaum [ A T ] yahoo.fr
date: 2006
desc: transform a genbank/XML file into html+svg
-->
<xsl:param name="screen-width">100</xsl:param>
<xsl:param name="seq-height">12</xsl:param>
<xsl:output
method="xml"
version="1.0"
encoding="UTF-8"
indent="yes"
/>
<xsl:template match="/">
<xul:window
id="findfile-window"
title="Find Files"
orient="horizontal"
xmlns="
http://www.mozilla.org/keymaster/gatekeeper/there.is.only.xul">
<xsl:apply-templates/>
</xul:window>
</xsl:template>
<xsl:template match="GBSet">
<xul:tabbox flex="1">
<xul:tabs >
<xsl:for-each select="GBSeq">
<xsl:element name="xul:tab">
<xsl:attribute name="id"><xsl:value-of select="generate-id(.)"/></xsl:attribute>
<xsl:attribute name="label"><xsl:value-of select="GBSeq_locus"/></xsl:attribute>
</xsl:element>
</xsl:for-each>
</xul:tabs>
<xul:tabpanels flex="1">
<xsl:for-each select="GBSeq">
<xul:tabpanel flex="1" id="searchpanel">
<xul:vbox flex="1">
<xul:hbox flex="1">
<xsl:apply-templates select="GBSeq_references"/>
</xul:hbox>
<xul:hbox flex="1">
<xul:iframe id="pubmed-iframe" src="about:blank" flex="1" style="overflow : auto; width : 30px; height : 300px; border:1px solid blue;"/>
<xul:box flex="1" style="overflow : auto; width : 30px; height : 300px; border:1px solid blue;">
<xsl:element name="svg:svg">
<xsl:attribute name="xul:flex">1</xsl:attribute>
<xsl:attribute name="width"><xsl:value-of select="2*$screen-width"/>+500</xsl:attribute>
<xsl:attribute name="height"><xsl:value-of select="count(GBSeq_feature-table/GBFeature)*20+5"/></xsl:attribute>
<xsl:attribute name="stroke">black</xsl:attribute>
<svg:g>
<svg:defs>
<svg:linearGradient x1="0%" y1="0%" x2="0%" y2="100%" id="metal">
<svg:stop offset="5%" stop-color="black" />
<svg:stop offset="50%" stop-color="whitesmoke" />
<svg:stop offset="95%" stop-color="black" />
</svg:linearGradient>
<svg:filter id="MyFilter" filterUnits="userSpaceOnUse" x="0%" y="0%" width="100%" height="100%">
<svg:feGaussianBlur in="SourceAlpha" stdDeviation="4" result="blur"/>
<svg:feOffset in="blur" dx="4" dy="4" result="offsetBlur"/>
<svg:feSpecularLighting in="blur" surfaceScale="5" specularConstant=".75"
specularExponent="20" lighting-color="lightgray"
result="specOut">
<svg:fePointLight x="-5000" y="-10000" z="20000"/>
</svg:feSpecularLighting>
<svg:feComposite in="specOut" in2="SourceAlpha" operator="in" result="specOut"/>
<svg:feComposite in="SourceGraphic" in2="specOut" operator="arithmetic"
k1="0" k2="1" k3="1" k4="0" result="litPaint"/>
<svg:feMerge>
<svg:feMergeNode in="offsetBlur"/>
<svg:feMergeNode in="litPaint"/>
</svg:feMerge>
</svg:filter>
</svg:defs>
<svg:g fill="red">
<xsl:call-template name="rect">
<xsl:with-param name="sequence-size" select="GBSeq_length" />
<xsl:with-param name="label" select="GBSeq_definition" />
<xsl:with-param name="x0" select="0" />
<xsl:with-param name="x1" select="GBSeq_length" />
</xsl:call-template>
<xsl:apply-templates select="GBSeq_feature-table" />
</svg:g>
</svg:g>
</xsl:element>
</xul:box>
</xul:hbox>
</xul:vbox>
</xul:tabpanel>
</xsl:for-each>
</xul:tabpanels>
</xul:tabbox>
</xsl:template>
<xsl:template match="GBSeq_references">
<xsl:element name="xul:tree">
<xsl:attribute name="flex">1</xsl:attribute>
<xsl:attribute name="enableColumnDrag">true</xsl:attribute>
<xsl:attribute name="onselect">var iframe= document.getElementById('pubmed-iframe');
if(iframe==null) { alert('cannot find iframe!'); return;}
switch(this.currentIndex+1)
{
<xsl:for-each select="GBReference">
case <xsl:value-of select="position()"/>:
iframe.setAttribute('src','
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&query_hl=1&list_uids=<xsl:value-of select="GBReference_pubmed"/>');
break;
</xsl:for-each>
default: iframe.setAttribute('src','about:blank');
break;
}
</xsl:attribute>
<xul:treecols>
<xul:treecol id="1" label="PMID" flex="1"/>
<xul:treecol id="3" label="Title" flex="1"/>
<xul:treecol id="4" label="Reference" flex="1"/>
<xul:treecol id="5" label="Authors" flex="1"/>
<xul:treecol id="6" label="Remarks" flex="2"/>
</xul:treecols>
<xul:treechildren >
<xsl:for-each select="GBReference">
<xul:treeitem>
<xul:treerow flex="1">
<xsl:element name="xul:treecell">
<xsl:attribute name="label">
<xsl:value-of select="GBReference_pubmed"/>
</xsl:attribute>
</xsl:element>
<xsl:element name="xul:treecell">
<xsl:attribute name="label">
<xsl:value-of select="GBReference_title"/>
</xsl:attribute>
</xsl:element>
<xsl:element name="xul:treecell">
<xsl:attribute name="label">
<xsl:value-of select="GBReference_journal"/>
</xsl:attribute>
</xsl:element>
<xsl:element name="xul:treecell">
<xsl:attribute name="label">
<xsl:for-each select="GBReference_authors/GBAuthor">
<xsl:value-of select="."/><xsl:text> </xsl:text>
</xsl:for-each>
</xsl:attribute>
</xsl:element>
<xsl:element name="xul:treecell">
<xsl:attribute name="label">
<xsl:value-of select="GBReference_remark"/>
</xsl:attribute>
</xsl:element>
</xul:treerow>
</xul:treeitem>
</xsl:for-each>
</xul:treechildren>
</xsl:element>
</xsl:template>
<xsl:template match="GBSeq_feature-table">
<xsl:apply-templates match="GBFeature" />
</xsl:template>
<xsl:template match="GBFeature">
<xsl:apply-templates select="GBFeature_intervals" />
</xsl:template>
<xsl:template match="GBFeature_intervals">
<xsl:apply-templates select="GBInterval" />
</xsl:template>
<xsl:template match="GBInterval">
<xsl:if test="GBInterval_from">
<xsl:call-template name="rect">
<xsl:with-param name="x0" select="GBInterval_from" />
<xsl:with-param name="x1" select="GBInterval_to" />
<xsl:with-param name="y" select="30+(count(../../preceding-sibling::*))*20" />
<xsl:with-param name="sequence-size" select="../../../../GBSeq_length" />
<xsl:with-param name="label" select="../../GBFeature_key" />
</xsl:call-template>
</xsl:if>
<xsl:if test="GBInterval_point">
<xsl:call-template name="rect">
<xsl:with-param name="x0" select="GBInterval_point" />
<xsl:with-param name="x1" select="GBInterval_point" />
<xsl:with-param name="y" select="30+(count(../../preceding-sibling::*))*20" />
<xsl:with-param name="sequence-size" select="../../../../GBSeq_length" />
<xsl:with-param name="label" select="../../GBFeature_key" />
</xsl:call-template>
</xsl:if>
</xsl:template>
<xsl:template name="rect">
<xsl:param name="x0" select="0" />
<xsl:param name="y" select="0" />
<xsl:param name="x1" select="0" />
<xsl:param name="label" />
<xsl:param name="height" select="10" />
<xsl:param name="sequence-size" />
<xsl:param name="i0" select="($x0 div $sequence-size)* $screen-width" />
<xsl:param name="width" select="(($x1 div $sequence-size)* $screen-width)-$i0" />
<xsl:param name="y2" select="$y+12" />
<xsl:if test="$i0 >= 0">
<xsl:element name="svg:rect">
<xsl:attribute name="x"><xsl:value-of select="$i0"/></xsl:attribute>
<xsl:attribute name="y"><xsl:value-of select="$y"/></xsl:attribute>
<xsl:attribute name="width"><xsl:value-of select="1+$width"/></xsl:attribute>
<xsl:attribute name="height"><xsl:value-of select="$height"/></xsl:attribute>
<xsl:attribute name="fill">url(#metal)</xsl:attribute>
<xsl:attribute name="stroke">black</xsl:attribute>
<xsl:attribute name="filter">url(#MyFilter)</xsl:attribute>
</xsl:element>
</xsl:if>
<xsl:element name="svg:text">
<xsl:attribute name="stroke">blue</xsl:attribute>
<xsl:attribute name="x"><xsl:value-of select="$screen-width+10"/></xsl:attribute>
<xsl:attribute name="y"><xsl:value-of select="$y2"/></xsl:attribute>
<xsl:value-of select="$x0"/> -><xsl:value-of select="$x1"/> : <xsl:value-of select="$label"/>
</xsl:element>
</xsl:template>
</xsl:stylesheet>